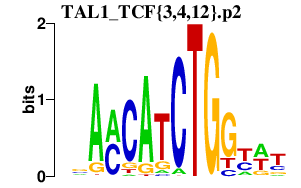
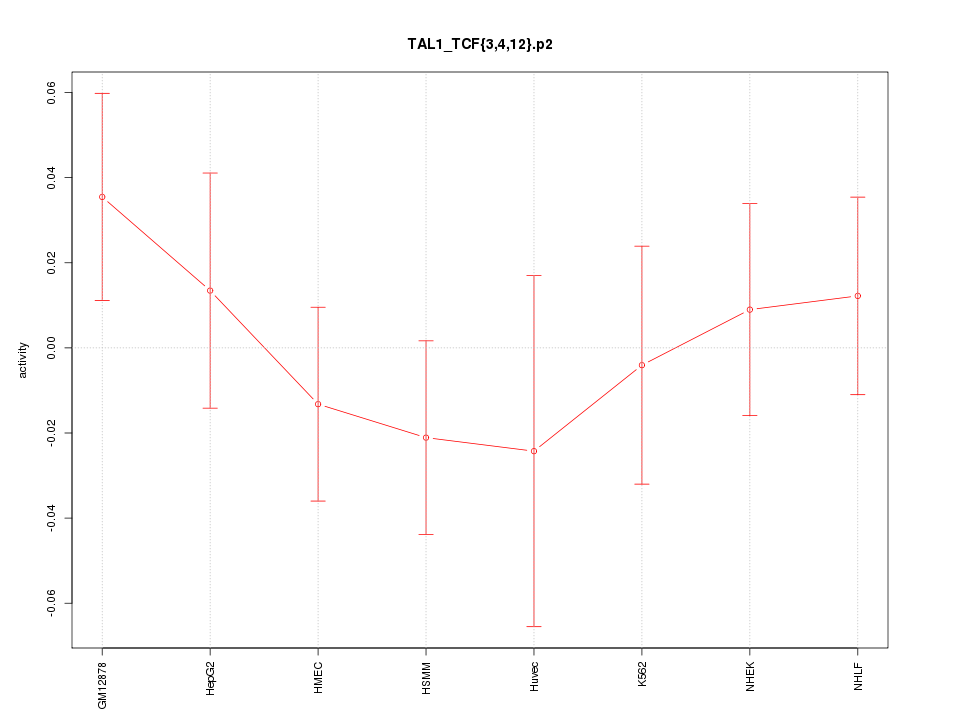
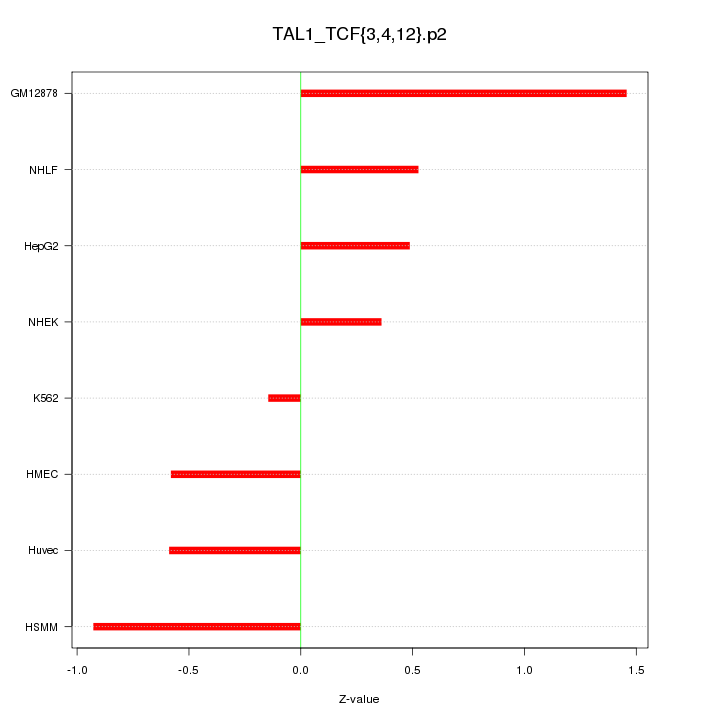
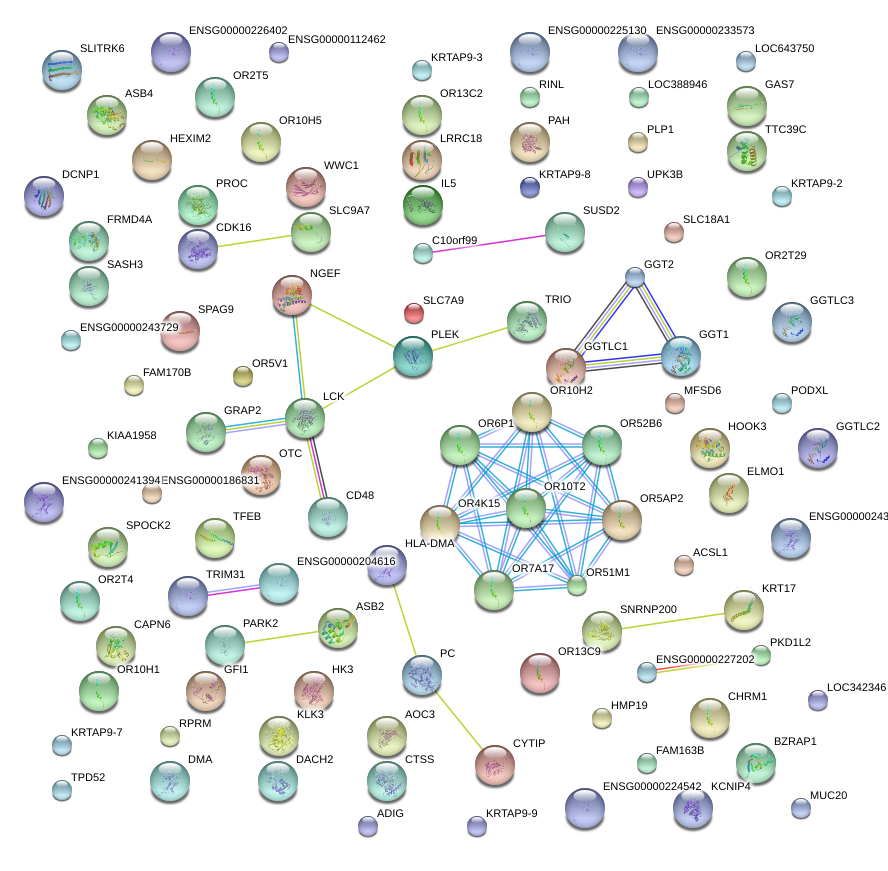
|
chr1_-_149004852
|
0.858
|
|
CTSS
|
cathepsin S
|
|
chr2_-_158008763
|
0.769
|
NM_004288
|
CYTIP
|
cytohesin 1 interacting protein
|
|
chr11_+_5367182
|
0.676
|
NM_001004756
|
OR51M1
|
olfactory receptor, family 51, subfamily M, member 1
|
|
chr17_+_38256710
|
0.667
|
NM_003734
|
AOC3
|
amine oxidase, copper containing 3 (vascular adhesion protein 1)
|
|
chr1_-_149004879
|
0.663
|
|
CTSS
|
cathepsin S
|
|
chr5_-_176258920
|
0.647
|
NM_002115
|
HK3
|
hexokinase 3 (white cell)
|
|
chr2_-_154043403
|
0.619
|
NM_019845
|
RPRM
|
reprimo, TP53 dependent G2 arrest mediator candidate
|
|
chr11_-_56166490
|
0.615
|
NM_001002925
|
OR5AP2
|
olfactory receptor, family 5, subfamily AP, member 2
|
|
chr9_-_106420305
|
0.593
|
NM_001001956
|
OR13C9
|
olfactory receptor, family 13, subfamily C, member 9
|
|
chr22_+_38652555
|
0.588
|
|
GRAP2
|
GRB2-related adaptor protein 2
|
|
chr1_+_246718487
|
0.527
|
NM_001004697
|
OR2T5
|
olfactory receptor, family 2, subfamily T, member 5
|
|
chr10_-_73518772
|
0.510
|
NM_001134434
NM_014767
|
SPOCK2
|
sparc/osteonectin, cwcv and kazal-like domains proteoglycan (testican) 2
|
|
chr17_+_40594046
|
0.507
|
NM_144608
|
HEXIM2
|
hexamthylene bis-acetamide inducible 2
|
|
chr1_+_32489426
|
0.503
|
NM_005356
|
LCK
|
lymphocyte-specific protein tyrosine kinase
|
|
chr10_-_14090459
|
0.499
|
|
FRMD4A
|
FERM domain containing 4A
|
|
chr13_-_85271329
|
0.483
|
NM_032229
|
SLITRK6
|
SLIT and NTRK-like family, member 6
|
|
chr1_-_156800017
|
0.445
|
NM_001160325
|
OR6P1
|
olfactory receptor, family 6, subfamily P, member 1
|
|
chr4_-_20594645
|
0.434
|
|
KCNIP4
|
Kv channel interacting protein 4
|
|
chr22_+_22907443
|
0.424
|
NM_019601
|
SUSD2
|
sushi domain containing 2
|
|
chr17_-_53761119
|
0.420
|
NM_004758
NM_024418
|
BZRAP1
|
benzodiazapine receptor (peripheral) associated protein 1
|
|
chr19_+_15699833
|
0.409
|
NM_013939
|
OR10H2
|
olfactory receptor, family 10, subfamily H, member 2
|
|
chr1_-_158948176
|
0.407
|
NM_001778
|
CD48
|
CD48 molecule
|
|
chr10_-_49792282
|
0.396
|
NM_001006939
|
LRRC18
|
leucine rich repeat containing 18
|
|
chr12_+_63510481
|
0.383
|
|
|
|
|
chr2_-_233586164
|
0.379
|
NM_019850
|
NGEF
|
neuronal guanine nucleotide exchange factor
|
|
chr5_-_131907071
|
0.377
|
NM_000879
|
IL5
|
interleukin 5 (colony-stimulating factor, eosinophil)
|
|
chr8_-_81155564
|
0.368
|
NM_001025252
|
TPD52
|
tumor protein D52
|
|
chr19_+_15765760
|
0.344
|
NM_001004466
|
OR10H5
|
olfactory receptor, family 10, subfamily H, member 5
|
|
chr1_-_246789396
|
0.334
|
NM_001004694
|
OR2T29
OR2T5
|
olfactory receptor, family 2, subfamily T, member 29
olfactory receptor, family 2, subfamily T, member 5
|
|
chr5_+_14493905
|
0.330
|
|
TRIO
|
triple functional domain (PTPRF interacting)
|
|
chr5_+_167651063
|
0.330
|
NM_001161661
NM_001161662
NM_015238
|
WWC1
|
WW and C2 domain containing 1
|
|
chrX_+_128741562
|
0.328
|
NM_018990
|
SASH3
|
SAM and SH3 domain containing 3
|
|
chr17_+_38256886
|
0.320
|
|
|
|
|
chr17_-_9863929
|
0.317
|
|
GAS7
|
growth arrest-specific 7
|
|
chr10_+_85923473
|
0.311
|
NM_207373
|
C10orf99
|
chromosome 10 open reading frame 99
|
|
chr6_-_163068788
|
0.304
|
|
PARK2
|
parkinson protein 2, E3 ubiquitin protein ligase (parkin)
|
|
chr12_-_101835026
|
0.300
|
|
PAH
|
phenylalanine hydroxylase
|
|
chr2_+_46560207
|
0.294
|
NM_001145051
|
LOC388946
|
transmembrane protein ENSP00000343375
|
|
chr6_-_41811916
|
0.291
|
NM_001167827
|
TFEB
|
transcription factor EB
|
|
chr7_+_94953148
|
0.288
|
NM_016116
NM_145872
|
ASB4
|
ankyrin repeat and SOCS box containing 4
|
|
chr6_-_29431950
|
0.275
|
NM_030876
|
OR5V1
|
olfactory receptor, family 5, subfamily V, member 1
|
|
chr17_+_36642240
|
0.273
|
NM_031962
|
KRTAP9-9
KRTAP9-3
|
keratin associated protein 9-9
keratin associated protein 9-3
|
|
chr3_+_196933481
|
0.272
|
|
MUC20
|
mucin 20, cell surface associated
|
|
chr19_-_44060729
|
0.271
|
NM_001195833
NM_198445
|
RINL
|
Ras and Rab interactor-like
|
|
chr2_+_191009265
|
0.266
|
|
MFSD6
|
major facilitator superfamily domain containing 6
|
|
chr14_-_93493519
|
0.265
|
NM_016150
|
ASB2
|
ankyrin repeat and SOCS box containing 2
|
|
chr6_-_30188845
|
0.255
|
NM_007028
|
TRIM31
|
tripartite motif containing 31
|
|
chr5_+_173405343
|
0.252
|
|
HMP19
|
HMP19 protein
|
|
chr7_+_75977680
|
0.252
|
NM_030570
NM_182684
|
UPK3B
|
uroplakin 3B
|
|
chr1_-_156635879
|
0.250
|
NM_001004475
|
OR10T2
|
olfactory receptor, family 10, subfamily T, member 2
|
|
chrX_+_102918409
|
0.248
|
NM_000533
NM_199478
|
PLP1
|
proteolipid protein 1
|
|
chr11_-_62445308
|
0.247
|
|
CHRM1
|
cholinergic receptor, muscarinic 1
|
|
chr18_+_19826734
|
0.244
|
NM_153211
|
TTC39C
|
tetratricopeptide repeat domain 39C
|
|
chr11_-_66431910
|
0.243
|
NM_022172
|
PC
|
pyruvate carboxylase
|
|
chr7_-_36992221
|
0.238
|
NM_130442
|
ELMO1
|
engulfment and cell motility 1
|
|
chrX_+_38096679
|
0.235
|
NM_000531
|
OTC
|
ornithine carbamoyltransferase
|
|
chr19_-_14853166
|
0.231
|
NM_030901
|
OR7A17
|
olfactory receptor, family 7, subfamily A, member 17
|
|
chr11_-_62445587
|
0.230
|
NM_000738
|
CHRM1
|
cholinergic receptor, muscarinic 1
|
|
chr5_+_173405298
|
0.230
|
NM_015980
|
HMP19
|
HMP19 protein
|
|
chr6_-_56515640
|
0.229
|
|
|
|
|
chrX_-_46503540
|
0.227
|
|
SLC9A7
|
solute carrier family 9 (sodium/hydrogen exchanger), member 7
|
|
chr11_-_71424614
|
0.225
|
|
|
|
|
chr20_+_36643251
|
0.221
|
NM_001018082
|
ADIG
|
adipogenin
|
|
chr17_+_45617676
|
0.220
|
|
|
|
|
chr14_+_19513442
|
0.216
|
NM_001005486
|
OR4K15
|
olfactory receptor, family 4, subfamily K, member 15
|
|
chr17_-_46479127
|
0.216
|
|
SPAG9
|
sperm associated antigen 9
|
|
chr4_-_185921384
|
0.213
|
|
ACSL1
|
acyl-CoA synthetase long-chain family member 1
|
|
chr2_-_96320185
|
0.212
|
|
SNRNP200
|
small nuclear ribonucleoprotein 200kDa (U5)
|
|
chr11_+_5558682
|
0.209
|
NM_001005162
|
OR52B6
|
olfactory receptor, family 52, subfamily B, member 6
|
|
chr16_+_4546491
|
0.209
|
NM_001145011
|
LOC342346
|
hypothetical protein LOC342346
|
|
chr22_+_22907417
|
0.207
|
|
SUSD2
|
sushi domain containing 2
|
|
chr9_+_114376709
|
0.206
|
|
KIAA1958
|
KIAA1958
|
|
chr8_-_20084892
|
0.206
|
NM_001135691
NM_001142324
NM_001142325
NM_003053
|
SLC18A1
|
solute carrier family 18 (vesicular monoamine), member 1
|
|
chrX_+_85404626
|
0.203
|
NM_001139515
|
DACH2
|
dachshund homolog 2 (Drosophila)
|
|
chrX_+_128741578
|
0.198
|
|
SASH3
|
SAM and SH3 domain containing 3
|
|
chrX_+_102918453
|
0.197
|
|
PLP1
|
proteolipid protein 1
|
|
chr9_-_135435165
|
0.196
|
NM_001080515
|
FAM163B
|
family with sequence similarity 163, member B
|
|
chr10_-_105410840
|
0.196
|
|
|
|
|
chrX_+_46967360
|
0.195
|
NM_001170460
|
CDK16
|
cyclin-dependent kinase 16
|
|
chr16_-_79811475
|
0.194
|
NM_001076780
NM_052892
|
PKD1L2
|
polycystic kidney disease 1-like 2
|
|
chr6_-_33028790
|
0.189
|
NM_006120
|
HLA-DMA
|
major histocompatibility complex, class II, DM alpha
|
|
chrX_-_110400406
|
0.186
|
NM_014289
|
CAPN6
|
calpain 6
|
|
chr10_-_50012058
|
0.185
|
NM_001164484
|
FAM170B
|
family with sequence similarity 170, member B
|
|
chr19_-_38052481
|
0.184
|
NM_001126335
NM_014270
|
SLC7A9
|
solute carrier family 7 (cationic amino acid transporter, y+ system), member 9
|
|
chr2_-_233586145
|
0.181
|
|
NGEF
|
neuronal guanine nucleotide exchange factor
|
|
chr2_+_68445821
|
0.178
|
NM_002664
|
PLEK
|
pleckstrin
|
|
chr1_-_92724181
|
0.175
|
NM_001127216
|
GFI1
|
growth factor independent 1 transcription repressor
|
|
chr5_-_134810696
|
0.169
|
NM_130848
|
C5orf20
|
chromosome 5 open reading frame 20
|
|
chr22_+_23309717
|
0.169
|
NM_013430
|
GGT1
|
gamma-glutamyltransferase 1
|
|
chr5_+_134331494
|
0.169
|
NM_178019
|
CATSPER3
|
cation channel, sperm associated 3
|
|
chr22_-_14829803
|
0.166
|
NM_001005239
|
OR11H1
|
olfactory receptor, family 11, subfamily H, member 1
|
|
chrX_+_102918089
|
0.161
|
NM_001128834
|
PLP1
|
proteolipid protein 1
|
|
chr17_+_72827227
|
0.160
|
|
SEPT9
|
septin 9
|
|
chr12_-_8110268
|
0.155
|
|
C3AR1
|
complement component 3a receptor 1
|
|
chr17_-_16816113
|
0.153
|
NM_012452
|
TNFRSF13B
|
tumor necrosis factor receptor superfamily, member 13B
|
|
chr9_+_12764988
|
0.151
|
NM_203403
|
C9orf150
|
chromosome 9 open reading frame 150
|
|
chr22_-_41672990
|
0.150
|
NM_007229
|
PACSIN2
|
protein kinase C and casein kinase substrate in neurons 2
|
|
chr1_-_169888373
|
0.150
|
NM_000261
|
MYOC
|
myocilin, trabecular meshwork inducible glucocorticoid response
|
|
chr12_-_7153068
|
0.148
|
NM_016546
|
C1RL
|
complement component 1, r subcomponent-like
|
|
chrX_-_15593074
|
0.147
|
NM_020665
|
TMEM27
|
transmembrane protein 27
|
|
chr22_+_34979118
|
0.141
|
|
APOL1
|
apolipoprotein L, 1
|
|
chr19_+_17499064
|
0.140
|
|
FAM129C
|
family with sequence similarity 129, member C
|
|
chr8_-_11966664
|
0.139
|
NM_001195257
NM_001037804
|
LOC100133267
DEFB130
|
defensin, beta 130-like
defensin, beta 130
|
|
chr10_+_71482362
|
0.138
|
NM_018649
|
H2AFY2
|
H2A histone family, member Y2
|
|
chr4_+_69465107
|
0.138
|
NM_014058
|
TMPRSS11E
|
transmembrane protease, serine 11E
|
|
chrX_+_125781427
|
0.137
|
NM_001122716
|
CXorf64
|
chromosome X open reading frame 64
|
|
chr16_+_11966464
|
0.136
|
NM_001192
|
TNFRSF17
|
tumor necrosis factor receptor superfamily, member 17
|
|
chr1_+_242283129
|
0.136
|
NM_006352
|
ZNF238
|
zinc finger protein 238
|
|
chr2_-_97775300
|
0.136
|
|
TMEM131
|
transmembrane protein 131
|
|
chr17_-_9864048
|
0.134
|
|
GAS7
|
growth arrest-specific 7
|
|
chr19_+_18069596
|
0.134
|
NM_015016
|
MAST3
|
microtubule associated serine/threonine kinase 3
|
|
chr14_-_20179689
|
0.133
|
NM_001001968
|
OR6S1
|
olfactory receptor, family 6, subfamily S, member 1
|
|
chr22_-_34886727
|
0.132
|
NM_145640
|
APOL3
|
apolipoprotein L, 3
|
|
chr12_-_101835510
|
0.128
|
NM_000277
|
PAH
|
phenylalanine hydroxylase
|
|
chr3_+_35697432
|
0.123
|
NM_198399
|
ARPP21
|
cAMP-regulated phosphoprotein, 21kDa
|
|
chr1_-_149004916
|
0.122
|
NM_004079
|
CTSS
|
cathepsin S
|
|
chr2_-_25320582
|
0.122
|
|
DNMT3A
|
DNA (cytosine-5-)-methyltransferase 3 alpha
|
|
chr6_-_163068749
|
0.121
|
|
PARK2
|
parkinson protein 2, E3 ubiquitin protein ligase (parkin)
|
|
chr6_-_169392771
|
0.120
|
|
|
|
|
chr10_-_105427898
|
0.119
|
|
SH3PXD2A
|
SH3 and PX domains 2A
|
|
chr17_+_71141108
|
0.119
|
NM_001162995
|
LOC643008
|
hypothetical protein LOC643008
|
|
chr13_-_26232777
|
0.118
|
NM_005288
|
GPR12
|
G protein-coupled receptor 12
|
|
chr6_+_29249289
|
0.118
|
NM_030905
|
OR2J2
|
olfactory receptor, family 2, subfamily J, member 2
|
|
chr10_+_11246934
|
0.118
|
NM_001025076
NM_001083591
|
CELF2
|
CUGBP, Elav-like family member 2
|
|
chr1_-_9052207
|
0.118
|
|
SLC2A5
|
solute carrier family 2 (facilitated glucose/fructose transporter), member 5
|
|
chr16_+_66836798
|
0.117
|
|
PLA2G15
|
phospholipase A2, group XV
|
|
chr16_-_70168449
|
0.117
|
NM_000353
|
TAT
|
tyrosine aminotransferase
|
|
chr17_+_19255083
|
0.116
|
NM_007148
|
RNF112
|
ring finger protein 112
|
|
chr19_+_56452775
|
0.115
|
NM_173635
|
C19orf75
|
chromosome 19 open reading frame 75
|
|
chr17_+_35372751
|
0.114
|
NM_178171
|
GSDMA
|
gasdermin A
|
|
chr12_-_8110196
|
0.114
|
NM_004054
|
C3AR1
|
complement component 3a receptor 1
|
|
chr1_+_246703249
|
0.114
|
NM_001005495
|
OR2T3
|
olfactory receptor, family 2, subfamily T, member 3
|
|
chr6_-_163068790
|
0.113
|
NM_004562
NM_013987
NM_013988
|
PARK2
|
parkinson protein 2, E3 ubiquitin protein ligase (parkin)
|
|
chr11_-_57752923
|
0.112
|
NM_001004471
|
OR10Q1
|
olfactory receptor, family 10, subfamily Q, member 1
|
|
chr10_-_121286034
|
0.112
|
NM_002925
|
RGS10
|
regulator of G-protein signaling 10
|
|
chrX_-_46503415
|
0.111
|
NM_032591
|
SLC9A7
|
solute carrier family 9 (sodium/hydrogen exchanger), member 7
|
|
chr1_+_245835478
|
0.111
|
NM_001001914
|
OR2G3
|
olfactory receptor, family 2, subfamily G, member 3
|
|
chr19_-_11549441
|
0.111
|
|
ACP5
|
acid phosphatase 5, tartrate resistant
|
|
chr12_-_14024187
|
0.110
|
NM_000834
|
GRIN2B
|
glutamate receptor, ionotropic, N-methyl D-aspartate 2B
|
|
chr19_-_11549484
|
0.109
|
|
ACP5
|
acid phosphatase 5, tartrate resistant
|
|
chrX_-_47941142
|
0.108
|
NM_021015
NM_175723
|
SSX5
|
synovial sarcoma, X breakpoint 5
|
|
chr1_-_23393808
|
0.108
|
NM_000864
|
HTR1D
|
5-hydroxytryptamine (serotonin) receptor 1D
|
|
chr7_+_155129943
|
0.106
|
NM_053043
|
RBM33
|
RNA binding motif protein 33
|
|
chr11_-_58368555
|
0.106
|
NM_145016
|
GLYATL2
|
glycine-N-acyltransferase-like 2
|
|
chr11_-_74478405
|
0.105
|
NM_001005285
|
OR2AT4
|
olfactory receptor, family 2, subfamily AT, member 4
|
|
chr7_+_31693155
|
0.105
|
NM_001145123
NM_006658
|
C7orf16
|
chromosome 7 open reading frame 16
|
|
chr7_+_113853790
|
0.103
|
NM_001172767
NM_148899
|
FOXP2
|
forkhead box P2
|
|
chr12_-_2814410
|
0.102
|
NM_031474
|
NRIP2
|
nuclear receptor interacting protein 2
|
|
chr9_+_115303527
|
0.101
|
NM_130795
|
RGS3
|
regulator of G-protein signaling 3
|
|
chr17_+_36515166
|
0.101
|
NM_001146041
|
KRTAP4-9
|
keratin associated protein 4-9
|
|
chr3_-_129668780
|
0.101
|
NM_153330
|
DNAJB8
|
DnaJ (Hsp40) homolog, subfamily B, member 8
|
|
chr11_-_55460451
|
0.101
|
NM_006637
|
OR5I1
|
olfactory receptor, family 5, subfamily I, member 1
|
|
chr17_+_44642587
|
0.098
|
NM_001135186
NM_016428
|
ABI3
|
ABI family, member 3
|
|
chr11_-_4902144
|
0.096
|
NM_001005237
|
OR51G1
|
olfactory receptor, family 51, subfamily G, member 1
|
|
chr3_+_49019498
|
0.096
|
NM_018031
|
WDR6
|
WD repeat domain 6
|
|
chr14_+_38804226
|
0.096
|
NM_203354
|
CTAGE5
|
CTAGE family, member 5
|
|
chr22_+_34979062
|
0.095
|
NM_001136540
NM_001136541
NM_003661
NM_145343
|
APOL1
|
apolipoprotein L, 1
|
|
chr2_-_240618518
|
0.095
|
NM_001005853
|
OR6B2
|
olfactory receptor, family 6, subfamily B, member 2
|
|
chr4_+_54790020
|
0.095
|
NM_006206
|
PDGFRA
|
platelet-derived growth factor receptor, alpha polypeptide
|
|
chr12_+_122510338
|
0.095
|
NM_180699
|
SNRNP35
|
small nuclear ribonucleoprotein 35kDa (U11/U12)
|
|
chr13_+_107719977
|
0.094
|
NM_001145645
NM_006573
|
TNFSF13B
|
tumor necrosis factor (ligand) superfamily, member 13b
|
|
chr2_-_25320299
|
0.093
|
|
DNMT3A
|
DNA (cytosine-5-)-methyltransferase 3 alpha
|
|
chr1_+_116181927
|
0.092
|
|
NHLH2
|
nescient helix loop helix 2
|
|
chr11_-_65182411
|
0.091
|
|
RELA
|
v-rel reticuloendotheliosis viral oncogene homolog A (avian)
|
|
chrX_-_111809929
|
0.090
|
NM_178175
|
LHFPL1
|
lipoma HMGIC fusion partner-like 1
|
|
chr7_+_155130086
|
0.090
|
|
RBM33
|
RNA binding motif protein 33
|
|
chr15_-_40236061
|
0.090
|
NM_213600
|
PLA2G4F
|
phospholipase A2, group IVF
|
|
chr6_+_167456230
|
0.089
|
NM_031409
|
CCR6
|
chemokine (C-C motif) receptor 6
|
|
chrX_-_19598912
|
0.089
|
NM_001184960
|
SH3KBP1
|
SH3-domain kinase binding protein 1
|
|
chr2_+_232165818
|
0.089
|
NM_152614
|
C2orf57
|
chromosome 2 open reading frame 57
|
|
chrX_-_110400321
|
0.089
|
|
CAPN6
|
calpain 6
|
|
chr5_-_142764295
|
0.088
|
|
NR3C1
|
nuclear receptor subfamily 3, group C, member 1 (glucocorticoid receptor)
|
|
chr8_-_86441115
|
0.087
|
NM_001164830
|
CA1
|
carbonic anhydrase I
|
|
chr6_+_126282078
|
0.087
|
|
NCOA7
|
nuclear receptor coactivator 7
|
|
chrX_+_48127911
|
0.087
|
NM_001034832
NM_001040612
NM_005636
NM_175729
|
SSX4B
SSX4
|
synovial sarcoma, X breakpoint 4B
synovial sarcoma, X breakpoint 4
|
|
chr7_+_149104063
|
0.087
|
NM_198455
|
SSPO
|
SCO-spondin homolog (Bos taurus)
|
|
chr16_+_2528022
|
0.086
|
|
PDPK1
|
3-phosphoinositide dependent protein kinase-1
|
|
chr22_-_38258716
|
0.084
|
NM_194326
|
RPS19BP1
|
ribosomal protein S19 binding protein 1
|
|
chr6_-_30236667
|
0.084
|
NM_006778
NM_052828
|
TRIM10
|
tripartite motif containing 10
|
|
chr19_-_11549508
|
0.082
|
NM_001611
|
ACP5
|
acid phosphatase 5, tartrate resistant
|
|
chr20_-_3635774
|
0.081
|
NM_023068
|
SIGLEC1
|
sialic acid binding Ig-like lectin 1, sialoadhesin
|
|
chr7_+_155129904
|
0.079
|
|
RBM33
|
RNA binding motif protein 33
|
|
chrX_+_41191630
|
0.079
|
NM_022567
|
NYX
|
nyctalopin
|
|
chr19_-_4289750
|
0.078
|
NM_001013841
NM_017720
|
STAP2
|
signal transducing adaptor family member 2
|
|
chr12_-_101835183
|
0.078
|
|
PAH
|
phenylalanine hydroxylase
|
|
chr10_+_7785347
|
0.077
|
|
ITIH2
|
inter-alpha (globulin) inhibitor H2
|
|
chr2_-_225519974
|
0.077
|
|
DOCK10
|
dedicator of cytokinesis 10
|
|
chr21_-_44506526
|
0.077
|
NM_013369
NM_175867
|
DNMT3L
|
DNA (cytosine-5-)-methyltransferase 3-like
|
|
chr3_-_197726587
|
0.077
|
NM_001077657
|
C3orf43
|
chromosome 3 open reading frame 43
|
|
chr17_-_1037312
|
0.076
|
NM_001159746
|
ABR
|
active BCR-related gene
|
|
chr12_+_118590039
|
0.075
|
NM_006253
|
PRKAB1
|
protein kinase, AMP-activated, beta 1 non-catalytic subunit
|
|
chr11_+_65164167
|
0.075
|
NM_153253
|
SIPA1
|
signal-induced proliferation-associated 1
|
|
chr5_-_180560535
|
0.074
|
NM_203297
|
TRIM7
|
tripartite motif containing 7
|
|
chr11_-_56914668
|
0.074
|
NM_002728
|
PRG2
|
proteoglycan 2, bone marrow (natural killer cell activator, eosinophil granule major basic protein)
|
|
chr6_-_41811323
|
0.074
|
|
TFEB
|
transcription factor EB
|
|
chr6_-_41230003
|
0.073
|
NM_178174
|
TREML1
|
triggering receptor expressed on myeloid cells-like 1
|
|
chr12_+_55674496
|
0.073
|
NM_007264
|
GPR182
|
G protein-coupled receptor 182
|
|
chr19_+_2340768
|
0.073
|
NM_182973
|
TMPRSS9
|
transmembrane protease, serine 9
|
|
chr12_+_50568010
|
0.073
|
NM_001130015
NM_182608
|
ANKRD33
|
ankyrin repeat domain 33
|
|
chr15_+_79276273
|
0.073
|
NM_001172128
|
IL16
|
interleukin 16 (lymphocyte chemoattractant factor)
|